S. Burrill, S. Elizalde, M. Mishna and L. Yen, A generating tree approach to k-nonnesting partitions and permutations,
Ann. Comb. 20 (2016) 453-485.
S. Burrill and L. Yen, Constructing skolem sequences via generating trees, arXiv:1301.6424.
W.Y.C. Chen, J. Qin and C.M. Reidys, Efficient counting and asymptotics of k-noncrossing tangled diagrams,
Electron. J. Combin. 16 (2009) Research Paper 37, 8 pp.
T. Feierl, Asymptotics for the number of walks in a weyl chamber of type B,
Random Structures Algorithms 45 (2014) 261-305.
T. Feierl, Random lattice walks in a weyl chamber of type A or B and non-intersecting lattice paths,
Ph.D. Thesis, University of Vienna, 2009.
H.S.W. Han and C.M. Reidys, The 5'-3' distance of RNA secondary structures, J. Comput. Biol. 19 (2012) 867-878.
H.S.W. Han and C.M. Reidys,
Pseudoknot RNA structures with arc-length 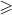 4,
J. Comput. Biol. 15 (2008) 1195-1208.
4,
J. Comput. Biol. 15 (2008) 1195-1208.
F.W.D. Huang, L.Y.M. Li and C.M. Reidys. Sequence-structure relations of pseudoknot RNA,
BMC bioinformatics 10 (2009) 1.
F.W.D. Huang, W.W.J. Peng and C.M. Reidys, Folding 3-noncrossing RNA pseudoknot structures,
J. Comput. Biol. 16 (2009) 1549-1575.
F.W.D. Huang, J. Qin, C.M. Reidys and P.F. Stadler, RNA-RNA interaction prediction: partition function and base pair pairing probabilities,
arXiv:0903.3283.
F.W.D. Huang and C.M. Reidys, Statistics of canonical RNA pseudoknot structures,
J. Comput. Biol. 253 (2008) 570-578.
F.W.D. Huang and C.M. Reidys, On the uniform generation of modular diagrams,
Electron. J. Combin. 17 (2010) Research Paper 175, 16 pp.
E.Y. Jin, J. Qin and C.M. Reidys, On k-noncrossing partitions, arXiv:0710.5014.
E.Y. Jin, J. Qin and C.M. Reidys, Neutral networks of sequence to shape maps,
Journal of Theoretical Biology 250 (2008) 484-497.
E.Y. Jin and C.M. Reidys, Combinatorial design of pseudoknot RNA,
Adv. in Appl. Math. 42 (2009) 135-151.
E.Y. Jin and C.M. Reidys, On the decomposition of k-noncrossing RNA structures,
Adv. in Appl. Math. 44 (2010) 53-70.
Z.C. Lin, Restricted inversion sequences and enhanced 3-noncrossing partitions, European J. Combin. 70 (2018) 202-211.
G. Ma and C.M. Reidys, Canonical RNA pseudoknot structures, J. Comput. Biol. 15 (2008) 1257-1273.
E. Marberg, Crossings and nestings in colored set partitions,
Electron. J. Combin. 20(4) (2013) Paper 6, 30 pp.
J. Marsh and S. Schroll, Trees, RNA secondary structures and cluster combinatorics, arXiv:1010.3763.
R. Müller and M.E. Nebel, Combinatorics of RNA secondary structures with base triples,
J. Comput. Biol. 22 (2015) 619-648.
J. Qin and C.M. Reidys, A combinatorial framework for RNA tertiary interaction, arXiv:0710.3523.
J. Qin and C.M. Reidys, Random 3-noncrossing partitions, arXiv:0910.2608.
J. Qin and C.M. Reidys, Random k-noncrossing partitions, arXiv:0911.2960.
C.M. Reidys, Local connectivity of neutral networks, Bull. Math. Biol. 71 (2009) 265-290.
C.M. Reidys, Combinatorial Computational Biology of RNA, Springer, New York, 2011.
S.H.F. Yan, Bijections for inversion sequences, ascent sequences and 3-nonnesting set partitions,
Appl. Math. Comput. 325 (2018) 24-30.